Download postgresql-8.3.5. ./configure --prefix=/Users/tjc/gmod/pgsl --with-pgport=2432 --with-includes=/Developer make make install cd /Users/tjc/gmod/pgsl bin/initdb -D data/ Added the line to data/postgresql.conf: listen_addresses = 'localhost' bin/postmaster -D data & bin/createuser --createdb tim bin/createlang plpgsql template1 bin/createdb --port=2432 test
Download stable release (gmod-1.0.tar.gz) Install BioPerl (http://www.bioperl.org/wiki/Installing_Bioperl_for_Unix) Install go-perl http://search.cpan.org/~cmungall/go-perl/ Install Bundle::GMOD from cpan setenv GMOD_ROOT /usr/local/gmod setenv CHADO_DB_NAME test setenv CHADO_DB_USERNAME tim setenv CHADO_DB_PORT 2432 Chado installation: perl Makefile.PL make sudo make install make load_schema make prepdb make ontologies
NC_001142:
You may find the following loading fails and you need to insert 'processed_transcript' as a sequence ontology cvterm. Or change them to 'mature_transcript' in the gff file (e.g. sed s'|processed_transcript|mature_transcript|' NC_001142.gff).
cat NC_001142.gbk | perl /usr/local/bin/bp_genbank2gff3.pl -noCDS -in stdin -out stdout > NC_001142.gff cat NC_001142.gff | perl /usr/local/bin/gmod_bulk_load_gff3.pl -dbname test -organism "Saccharomyces cerevisiae" -dbuser tim -dbport 2432 -dbpass dd -recreate_cache
NC_008783:
Add this to the organism table:
INSERT INTO organism
(abbreviation, genus, species, common_name, organism_id) VALUES
('B.bacilliformis', 'Bartonella', 'bacilliformis', 'BB', 8783);
cat NC_008783.gbk | perl /usr/local/bin/bp_genbank2gff3.pl -noCDS -in stdin -out stdout > NC_008783.gff
cat NC_008783.gff | perl /usr/local/bin/gmod_bulk_load_gff3.pl -dbname test -organism "BB" -dbuser tim -dbport 2432 -dbpass dd -recreate_cache
Artemis does not (by default) look for sequences that are loaded as a 'region'.
So change the sequence type of NC_001142 from region to a chromosome:
UPDATE feature SET
type_id=(select cvterm_id from cvterm where cv_id=(
select cv_id from cv where name='sequence') and name='chromosome')
WHERE uniquename='NC_008783';
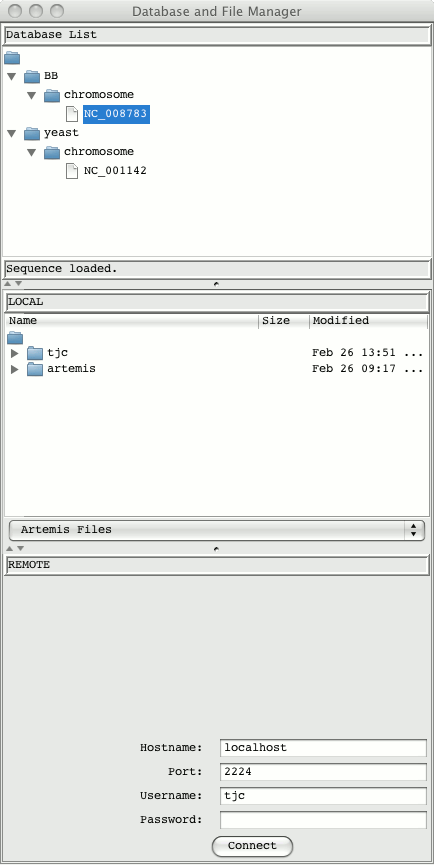
./art -Dchado="localhost:2432/test?tim" -Dibatis \
-Djdbc.drivers=org.postgresql.Driver
If this is successful then the login window will appear and the database manager will then open: